Combining PNM and FIX regressors in a FEAT analysis
This page describes a process for combining noise regressors identified by tools such as PNM and ICA / FIX, and preparing them for use within a single FEAT model, thus allowing the user to clean their data in a single step.
Note that it is strongly recommended to combine regressors from tools like PNM and FIX into a single regression step, rather than performing them separately. The reason for this is that variance can be reintroduced into the "cleaned" data if regressions are performed separately.
Step 1: Pre-processing
Before combining your sources of noise you should follow the relevant instructions for PNM and MELODIC. ICA noise components can be manually classified using FSLeyes, or automatically classified using a tool such as FIX or ICA-AROMA. Other pre-processing steps such as motion correction, registration and smoothing should be carried out normally.
Your aim is have ready all necessary files - voxelwise confounds generated with PNM, and the time series of noise components identified via MELODIC and FIX, along with any other noise time series information you may wish to include in your FEAT model.
Step 2: Extracting time series for noise ICs
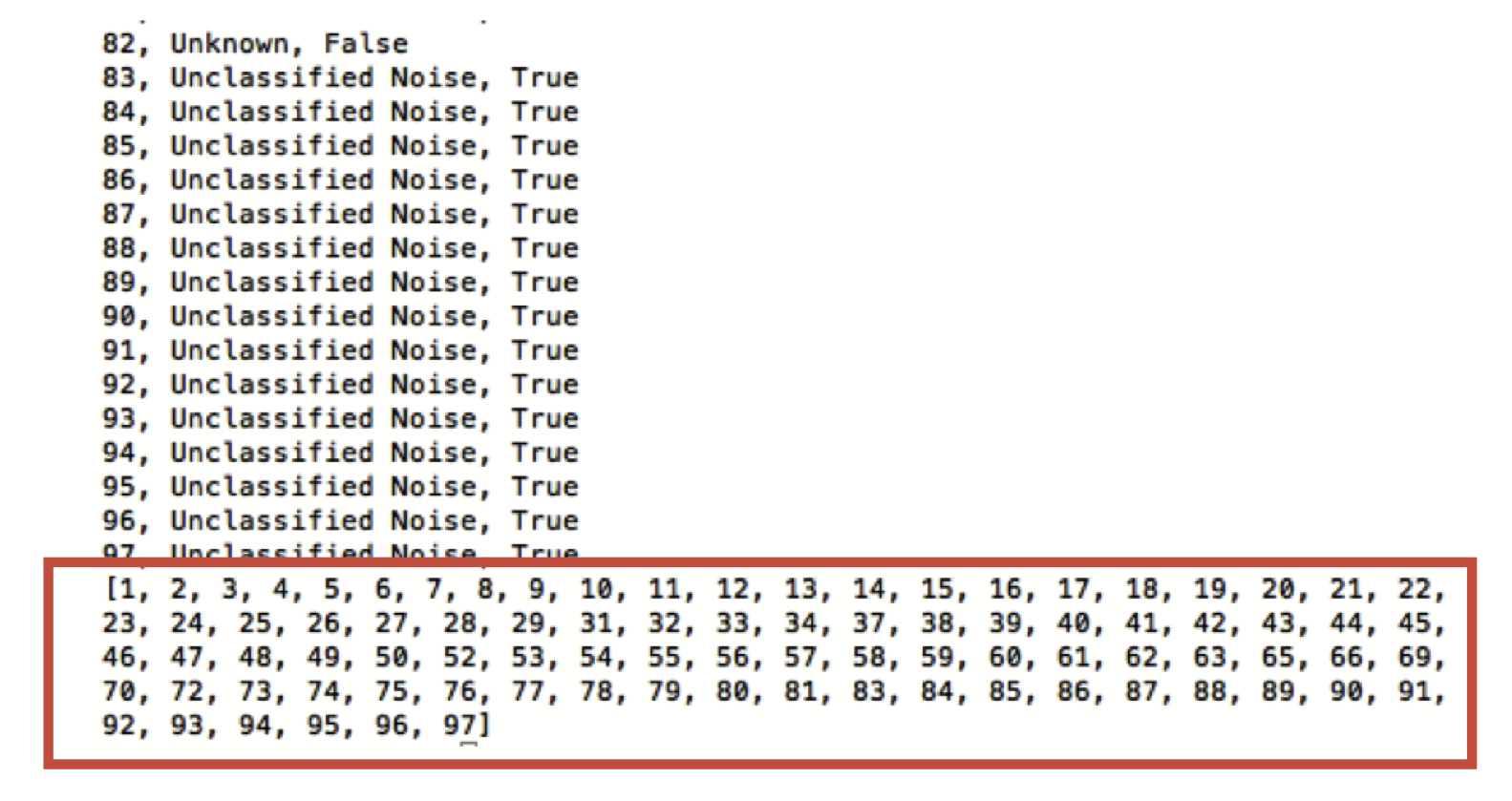
After using FSLeyes or FIX to classify your MELODIC ICs as either signal or noise, you will have a file which contains the labels for each IC. The final line of this file contains the numbers of all noise components.
These numbers can be used to index the file containing the time-series information for each component - this is produced by MELODIC, and is named melodic_Tmodes, located within the filtered_func_data.ica folder of your MELODIC directory. To regress these noise components from your fMRI data, you need to generate a file that contains only the noise component timecourses from melodic_Tmodes. This new noise time series text file (named for example: ICA_noise.txt) should be space separated. In addition to ICA identified noise components, signal associated with white matter, CSF or motion outliers can also be appended to this text file.
The fsl_ents command-line tool can extract the time series of noise components from a MELODIC directory and prepare a file suitable for using as a confound EV file within FEAT. For example, if your label file is called fix4melview_training_thr20.txt, you can create a text file which contains the time series of all noise components by running:
You may also pass additional confound files to fsl_ents - for example, if you have useed fsl_motion_outliers on your data to identify outlier time points (saving its output to outliers.txt), you can pass it to fsl_ents to produce a single file that contains both the time series of noise ICs, and outlier timepoint regressors:
Step 3: Inputting the noise text files into your model
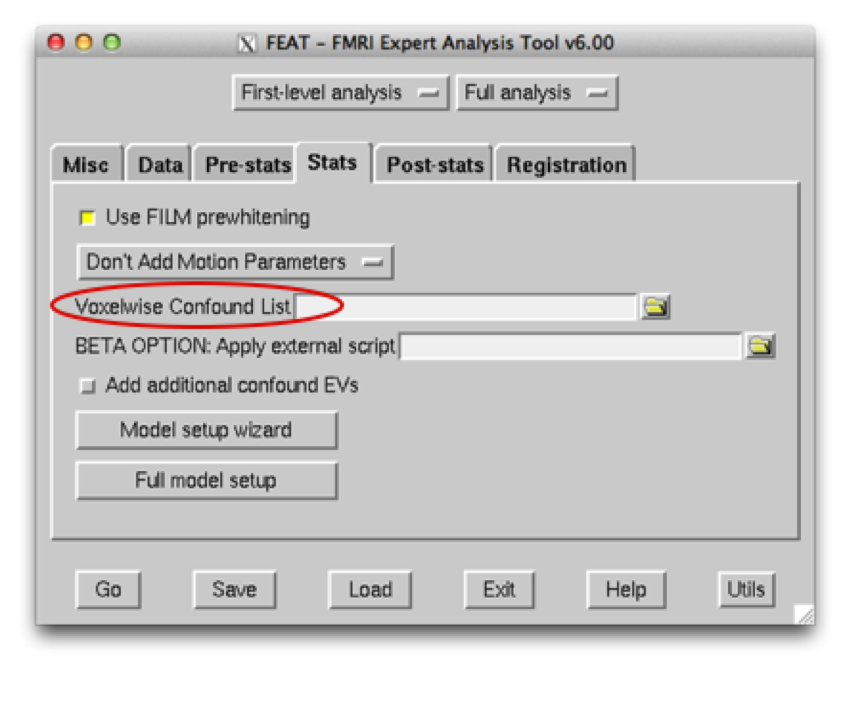
If you have carried out both PNM and ICA, you should now have two text files – one for your PNM noise output, which should be input into the Voxel Confound List in FEAT:
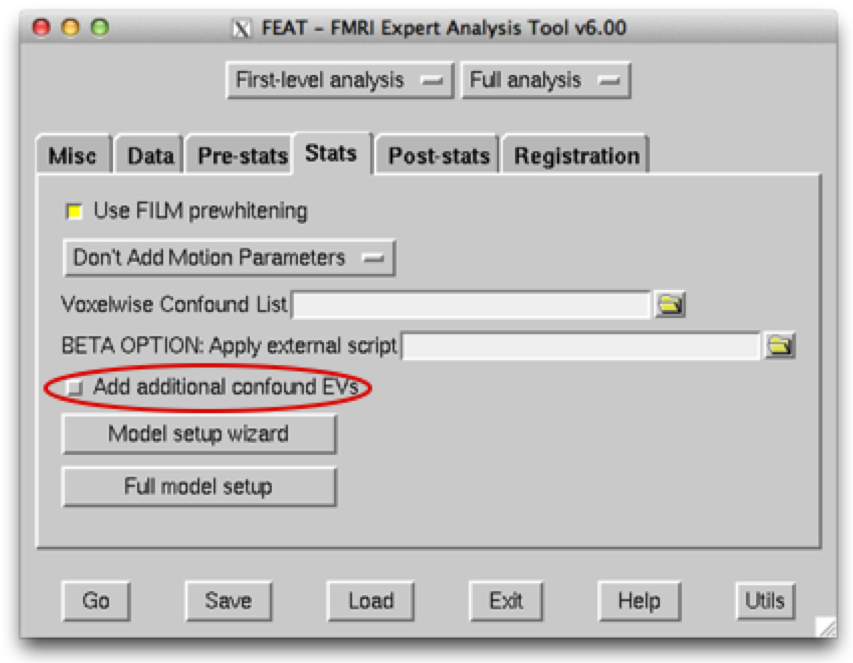
And another text file containing the time series information of ICA noise components, along with any other sources of noise that you wish to model. This file should be space delimited and should be input into the Add additional confound EVs box. The input within the Data tab should be your raw fMRI data - FEAT can then be run from scratch, including all pre-processing steps.
The model
The PNM noise text file and FIX noise text file will be combined by FEAT and input into your model as one long series of regressors of no interest. Many of these regressors are likely to not be independent and therefore we do not expect the degrees of freedom to be overly reduced.
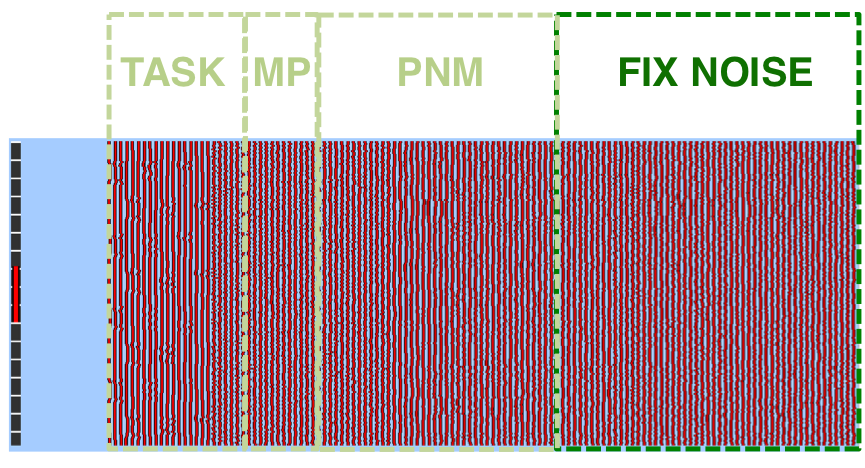
If you are working with resting state fMRI data, there will be no task EVs in your FEAT model. In this case, the res4d.nii.gz file obtained from running FEAT as described above will be your "cleaned" data, ready to be used in your resting state analysis pipeline.
Referencing
If you are using the tools and methods described here in your research, please select the appropriate publications from the list below pertaining to FIX and PNM for referencing.
G. Salimi-Khorshidi, G. Douaud, C.F. Beckmann, M.F. Glasser, L. Griffanti S.M. Smith. Automatic denoising of functional MRI data: Combining independent component analysis and hierarchical fusion of classifiers. NeuroImage, 90:449-68, 2014
L. Griffanti, G. Salimi-Khorshidi, C.F. Beckmann, E.J. Auerbach, G. Douaud, C.E. Sexton, E. Zsoldos, K. Ebmeier, N. Filippini, C.E. Mackay, S. Moeller, J.G. Xu, E. Yacoub, G. Baselli, K. Ugurbil, K.L. Miller, and S.M. Smith. ICA-based artefact removal and accelerated fMRI acquisition for improved resting state network imaging. NeuroImage, 95:232-47, 2014
Brooks JC, Beckmann CF, Miller KL, Wise RG, Porro CA, Tracey I, Jenkinson M. Physiological noise modelling for spinal functional magnetic resonance imaging studies. Neuroimage, 39(2):680-92, 2008.
Harvey AK, Pattinson KT, Brooks JC, Mayhew SD, Jenkinson M, Wise RG. Brainstem functional magnetic resonance imaging: disentangling signal from physiological noise. J Magn Reson Imaging. 28(6):1337-44, 2008.
Birn RM, Diamond JB, Smith MA, Bandettini PA. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage, 31(4):1536-48, 2006.
Glover GH, Li TQ, Ress D. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn Reson Med., 44(1):162-7, 2000.
Shmueli K, van Gelderen P, de Zwart JA, Horovitz SG, Fukunaga M, Jansma JM, Duyn JH. Low-frequency fluctuations in the cardiac rate as a source of variance in the resting-state fMRI BOLD signal. Neuroimage, 38(2):306-20, 2007.
Chang C, Cunningham JP, Glover GH. Influence of heart rate on the BOLD signal: the cardiac response function. Neuroimage, 44(3):857-69, 2009.
Cohen-Adad J, Gauthier CJ, Brooks JC, Slessarev M, Han J, Fisher JA, Rossignol S, Hoge RD. BOLD signal responses to controlled hypercapnia in human spinal cord. Neuroimage, 50(3):1074-84, 2010.