FDT utilities
Vecreg - registration of vector images
After running dtifit or bedpostx, it is often useful to register vector data to another space. For example, one might want to represent V1 for different subjects in standard space. vecreg is a command line tool that allows to perform such registration.
Vector images cannot be registered by simply applying a transformation (as calculated by, say, FLIRT) to every voxel's coordinates. The corresponding vectors have to be reoriented accordingly (see D. Alexander 2001, IEEE-TMI 20:1131-39).
vecreg performs this operation for you. The image below shows the effect of applying vecreg (right) to the V1 image on the left, compared to simply applying voxelwise transformation (e.g. using applyxfm4D) to the vectors (centre).
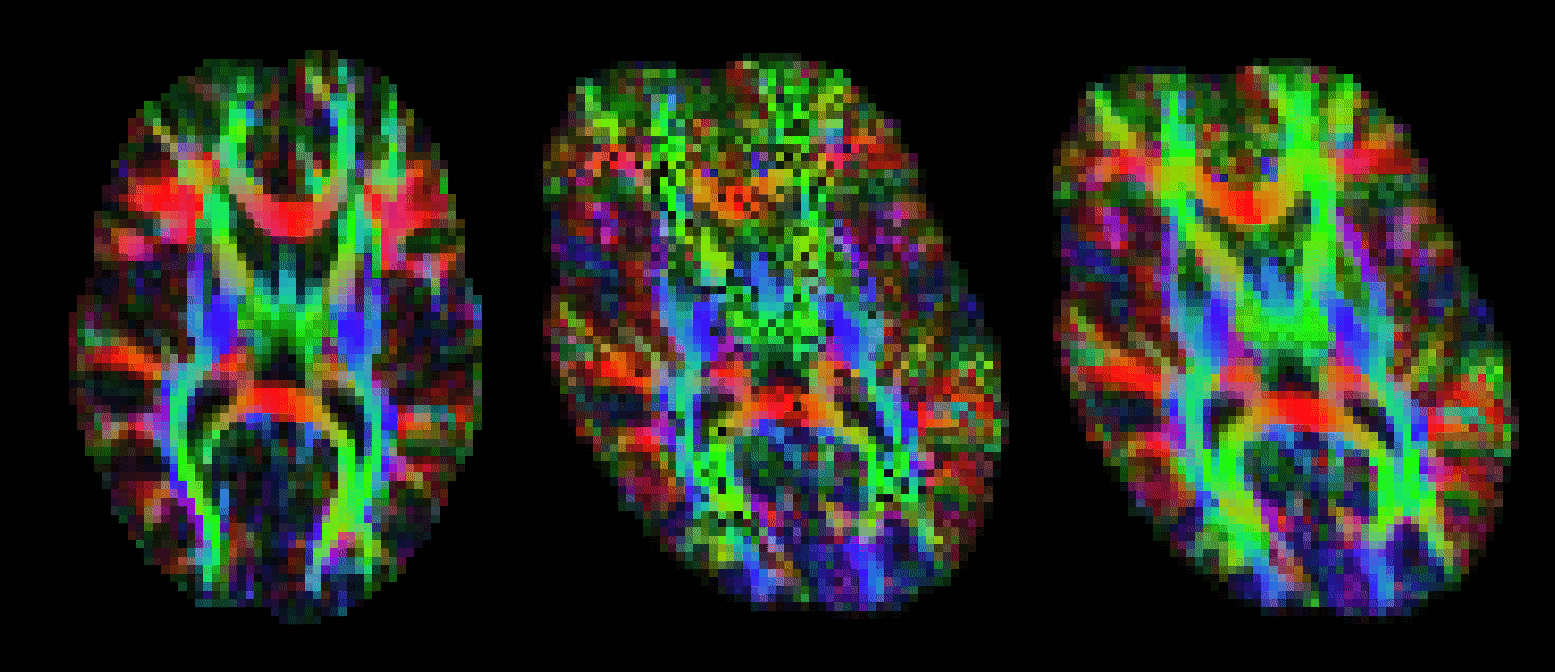
Info
vecreg does not calculate a transformation, but simply applies a given transformation to the input vector field. vecreg can apply a linear transformation calculated with FLIRT, or a non-linear transformation calculated by FNIRT.
Types of input that may be used for vecreg from DTIFIT: V1,V2,V3,tensor from BEDPOSTX: dyads1, dyads2, etc.
Command line options
vecreg -i <input4D> -o <output4D> -r <refvol> [-t <transform>]
Compulsory arguments (You MUST set one or more of):
-i,--input filename for input vector or tensor field
-o,--output filename for output registered vector or tensor field
-r,--ref filename for reference (target) volume
Optional arguments (You may optionally specify one or more of):
-v,--verbose switch on diagnostic messages
-h,--help display this message
-t,--affine filename for affine transformation matrix
-w,--warpfield filename for 4D warp field for nonlinear registration
--rotmat filename for secondary affine matrix
if set, this will be used for the rotation of the vector/tensor field
--rotwarp filename for secondary warp field
if set, this will be used for the rotation of the vector/tensor field
--interp interpolation method : nearestneighbour, trilinear (default), sinc or spline
-m,--mask brain mask in input space
--refmask brain mask in output space (useful for speed up of nonlinear reg)
Qboot
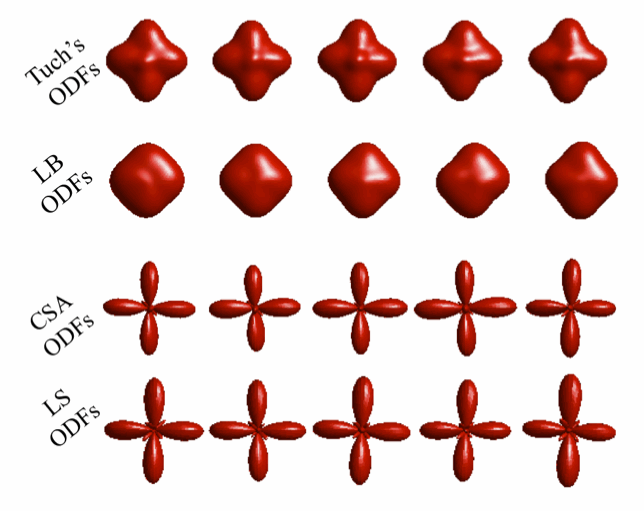
qboot is a command line tool that allows estimation of diffusion ODFs and fibre orientations from them. Its output can be used as an input for probtrackx in order to perform probabilistic tractography.
ODF estimation is performed using a real spherical harmonics basis. Fibre orientations are estimated as the local maxima of the ODFs. Both deterministic and probabilistic estimation can be performed. For the latter, residual bootstrap is performed to infer on the ODF shape and obtain a distribution of fibre orientations. For more details on the implementation see Sotiropoulos2011 (S.N. Sotiropoulos, I. Aganj, S. Jbabdi, G. Sapiro, C. Lenglet and T.E. Behrens, "Inference on Constant Solid Angle Orientation Distribution Functions from Diffusion-Weighted MRI", p.609, Quebec, Canada, OHBM, 2011).
qboot allows reconstruction of q-ball ODFs (Tuch DS, MRM 2004), CSA ODFs (Aganj I et al, MRM, 2010) and variants of them, obtained via Laplacian sharpening and Laplace-Beltrami regularization (Descoteaux et al, MRM, 2007). Both spherical harmonic coefficients of the reconstructed ODFs and fibre orientation estimates may be returned as output. A real spherical harmonic basis is employed (Aganj I et al, MRM, 2010).
Input files for qboot : Similar to dtifit and bedpostx, qboot needs a 4D data file, a binary mask_file, a bvecs and a bvals file.
Usage
qboot --help (for list of options)
Compulsory arguments (You MUST set one or more of):
-k,--data Data file
-m,--mask Mask file
-r,--bvecs b vectors file
-b,--bvals b values file
Optional arguments (You may optionally specify one or more of):
--ld,--logdir Output directory (default is logdir)
--forcedir Use the actual directory name given - i.e. don't add + to make a new directory
--q File provided with multi-shell data. Indicates the number of directions for each shell
--model Which model to use. 1=Tuch's ODFs, 2=CSA ODFs (default), 3=multi-shell CSA ODFs
--lmax Maximum spherical harmonic oder employed (must be even, default=4)
--npeaks Maximum number of ODF peaks to be detected (default 2)
--thr Minimum threshold for a local maxima to be considered an ODF peak. Expressed as a fraction of the maximum ODF value (default 0.4)
--ns,--nsamples Number of bootstrap samples (default is 50)
--lambda Laplace-Beltrami regularization parameter (default is 0)
--delta Signal attenuation regularization parameter for models=2,3 (default is 0.01)
--alpha Laplacian sharpening parameter for model=1 (default is 0, should be smaller than 1)
--seed Seed for pseudo-random number generator
--gfa Compute a generalised FA, using the mean ODF in each voxel
--savecoeff Save the ODF coefficients instead of the peaks. WARNING: These can be huge files, please use a few bootstrap samples and a low lmax!
--savemeancoeff Save the mean ODF coefficients across all samples
-V,--verbose Switch on diagnostic messages
-h,--help Display this message