BEDPOSTX
BEDPOSTX stands for Bayesian Estimation of Diffusion Parameters Obtained using Sampling Techniques. The X stands for modelling Crossing Fibres. bedpostx runs Markov Chain Monte Carlo sampling to build up distributions on diffusion parameters at each voxel. It creates all the files necessary for running probabilistic tractography. For an overview of the modelling carried out within bedpostx see this technical report.
bedpostx models crossing fibres within each voxel of the brain. Crucially, bedpostx automatically determines the number of crossing fibres per voxel. For details on the basic crossing fibre model see Behrens et al, NeuroImage 2007. By default, bedpostx runs a slightly more complex model Jbabdi et al. MRM 2012, where the diffusion coefficient is modelled using a Gamma distribution.
bedpostx takes about 15 hours to run on a single CPU, but will automatically batch if run on an SGE-capable system. There is also a GPU implementation called bedpostx_gpu which is about 100x faster.
Note that bedpostx is a wrapper script for a command-line tool called xfibres.
The remainder of this page is organised into the following sections:
BedpostX from the command line
Usage: bedpostx <subject directory> [options]
expects to find bvals and bvecs in subject directory
expects to find data and nodif_brain_mask in subject directory
expects to find grad_dev in subject directory, if -g is set options (old syntax)
-n (number of fibres per voxel, default 3)
-w (ARD weight, more weight means less secondary fibres per voxel, default 1)
-b (burnin period, default 1000)
-j (number of jumps, default 1250)
-s (sample every, default 25)
-model (Deconvolution model. 1: with sticks, 2: with sticks with a range of diffusivities (default), 3: with zeppelins)
-g (consider gradient nonlinearities, default off)
ALTERNATIVELY: you can pass on xfibres options onto directly bedpostx
For example: bedpostx <subject directory> --noard --cnonlinear
Type 'xfibres --help' for a list of available options
Default options will be bedpostx default (see above), and not xfibres default.
Note: Use EITHER old OR new syntax.
BedpostX from the GUI
In the FDT GUI, use the top left drop down menu to select BEDPOSTX.
-
Input directory: Use the browse button to select an input directory. That directory must contain the following files:
-
data.nii.gz: A 4D series of data volumes. This will include diffusion-weighted volumes and volume(s) with no diffusion weighting. -
nodif_brain_mask.nii.gz: 3D binary brain mask volume derived from running bet on nodif (i.e. on a volume with no diffusion weighting). -
bvecs(with no file extension): An ASCII text file containing a list of gradient directions applied during diffusion weighted volumes. -
bvals(with no file extension): An ASCII text file containing a list of bvalues applied during each volume acquisition.
Tip
Run bedpostx_datacheck in command line to check if your input directory contains the correct files required for bedpostx.
Outputs of BedpostX
bedpostx creates a new directory at the same level as the input directory called <indir>.bedpostX which contains all the files you need for probabilistic tractography. Highlights are (<i> indicates the i-th fibre. It ranges from 1 to the maximum number of fibres set in the advanced options.):
-
merged_th<i>samples- 4D volume - Samples from the distribution on theta -
merged_ph<i>samples- 4D volume - Samples from the distribution on phi
theta and phi together represent the principal diffusion direction in spherical polar co-ordinates
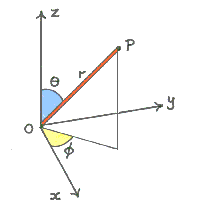
-
merged_f<i>samples- 4D volume - Samples from the distribution on anisotropic volume fraction (see technical report). -
mean_th<i>samples- 3D Volume - Mean of distribution on theta -
mean_ph<i>samples- 3D Volume - Mean of distribution on phi -
mean_f<i>samples- 3D Volume - Mean of distribution on f anisotropy
Note that in each voxel, fibres are ordered according to a decreasing mean f-value
-
mean_dsamples- 3D Volume - Mean of distribution on diffusivity d -
mean_d_stdsamples - 3D Volume - Mean of distribution on diffusivity variance parameter d_std (not produced if --model=1) -
mean_S0samples- 3D Volume - Mean of distribution on T2w baseline signal intensity S0 -
dyads<i>- Mean of PDD distribution in vector form. Note that this file can be loaded into FSLeyes for easy viewing of diffusion directions -
dyads<i>_dispersion- 3D Volume - Uncertainty on the estimated fibre orientation. Characterizes how wide the orientation distribution is around the respective PDD.(how is this calculated?) -
nodif_brain_mask- binary mask created from nodif_brain - copied from input directory
Advanced Options
You may change some options before running bedpostx, depending on the questions you want to ask or the quality of your diffusion data. The default values of these parameters are the ones used in the corresponding paper (Behrens et al, NeuroImage 2007).
- Fibres: Number of fibres modelled per voxel.
- Weight: Multiplicative factor for the prior on the additional modelled fibres. A smaller factor means more weighting for the additional fibres.
- Burnin: Number of iterations before starting the sampling. These might be increased if the data are noisy, and the MCMC needs more iterations to converge.
Additionally, the following alternative models are available in the advanced options:
-
Single-Shell Model : Use this option if the data contains only a single non-zero bvalue. This will revert the model to use the one described in Behrens et al, NeuroImage 2007 rather than in Jbabdi et al. MRM 2012 (i.e., the the diffusion coefficient is modelled as a single value rather than using a Gamma distribution). Command line argument:
--model=1. -
Model Noise Floor : Use this option if (part of) the data is operating close to the noise floor. This will fit one extra parameter f0 that captures the noise floor. This is also described in Jbabdi et al. MRM 2012. This option will output the posterior mean of the noise floor parameter mean_f0samples. Command line argument:
--f0 --ardf0. -
Rician Noise : Use this option to replace the default Gaussian noise assumption with Rician noise. This will output the posterior mean of a noise precision (i.e. inverse of variance) parameter
mean_tausamples. Command line argument:--rician.