Frequently asked questions about eddy
What are the GPU requirements?
You are probably here because you know that in the past there was a need to match the Cuda version of eddy with the version of the Cuda library that was installed on one's machine. That is no longer neccessary.
Now one only has to ensure that the currently installed drivers are 10.2 or later (10.2 is now almost 5 years old, so it is unlikely that your drivers are older than that). It also means that it has native support for architectures Kepler to Turing, and software emulation support for architectures later than Turing. People who have a very old GPU (Tesla or Fermi) will need to build eddy for an older version of Cuda. People who have a "very new" architecture GPU (Ampere or later) can build eddy for a newer version of Cuda. But I don't think it will make any/much difference to execution speed since I haven't used any of the new coding features.
How do I know what to put into my --acqp file?
The --acqp file is of the same format as the --datain (though one can now use --acqp also in topup) parameter for topup, and most likely you can re-use the one you created for topup. It is used to inform eddy what direction the distortions are likely to go in. Each row consists of a vector (three values) that specify what the phase-encode (PE) axis is and also what direction along that axis that imply higher frequency. So for example the two lines
The fourth element in each row is the time (in seconds) between reading the center of the first echo and reading the center of the last echo. It is the "dwell time" multiplied by "number of PE steps - 1" and it is also the reciprocal of the PE bandwidth/pixel.
If you have used a reasonably recent version of dcm2niix the conversion of each "run" (a set of images acquired after a single press "Go" on the scanner) will produce, in addition to the image file/files, a .json file. That .json file will have a "name-value" pair with the name "TotalReadoutTime" which will give you the value to use as the fourth element.
In the same .json file there will also be a field with the name "PhaseEncodingDirection", which will contain information relevant to the first three values. The following mapping will "often" be correct, but it will, annoyingly, also depend on the image transform that is stored in the image header.
| "PhaseEncodingDirection" | Phase-encode vector | "i" | [1 0 0] |
|---|---|
| "i-" | [-1 0 0] |
| "j" | [0 1 0] |
| "j-" | [0 -1 0] |
So, for example "PhaseEncodingDirection": "i" would suggest a phase-encode vector of [1 0 0]
Does it matter what I put into my --acqp file?
Almost not at all, as long as the same file is used for both topup and eddy.
Let us for example say that the "true" file should be
but that we happen to use one that looks like instead.Surely that must be disastrous?
No, in fact the results would be (almost) indistinguishable from each other.
The sign-swap for the PE-directions would mean that topup would sign-swap the entire field so that areas with higher field will look like they have a lower field, and vice versa. The twice as large "readout time" means that topup will underestimate the field by a factor 2. Hence, the estimated field will be sign-swapped and underestimated, which on the surface sounds like a bad thing.
But now we enter that field into eddy with the --topup parameter and also the "faulty" --acqp file. The sign-swap in the field and in the --acqp file will now cancel out by being multiplied together, so the displacements will still go in the correct direction. Similarily, the field is underestimated by a factor 2, but that is counteracted by a twice as large "sensitivity" to off-resonance indicated by the too large readout time. So the magnitude of the displacements is also correct.
So, does it never matter at all what I put into my --acqp file?
- at all
Let me start with the latter, at all. With the exception of the cases described below, it almost doesn't matter at all. If you use a total readout time that is an order of magnitude, or more, wrong that results will be "a little" different. That is because the regularisation is applied to the Hz-field, which means that if that is completely wrong the regularisation might be too strong or too lenient. But if you use a value in the range 0.005--0.5 seconds you should be fine. And all "true" values will definitely be in that range. But as I said before, if in doubt just use 0.05. - There is an additional little detail to the above. If you run
topupon two images where one of them are undistorted (for example if you use Synb0), then you can, and should, use a very small (1e-6) total readout time for the undistorted image. In that case it will not affect the scaling to Hz, and hence not the regularisation. - never
There are some special cases where it matters to get the --acqp file right, but unless you know exactly what you are doing it is generally best to avoid those cases. They would be: - If you acquire data with PE direction along two different axes (i.e. the x- and y-axis). In that case you need to get the signs right for the columns indicating the PE. But you can always use trial and error to find the correct combination.
- If you acquire data with different readout times. In that case you need at the very least to get the ratio between the times right.
- If you use a non-topup derived fieldmap, such as for example a dual echo-time gradient echo fieldmap, that you feed into eddy as the --field parameter. In this case you need to get all signs and times right, both when creating the field (for example using prelude) and when specifying its use in eddy through the --acqp.
I don't have a .json file. What do I put in my --acqp file?
There is the easy way, and then there is the very easy way. Let's start with the latter.
- The very easy way:
Run a movie (for example using fslview or FSLeyes) of the 4D file you plan to enter into topup.
Does the brain jump up and down?
If so, use
If so, use Is the brain essentially still?
If so, there has probably been a mistake in the acquisition and both images have been acquired with the same phase-encoding. In that case you cannot use topup on these data (though see Synb0 for a possible way to "rescue" the data). - The easy way:
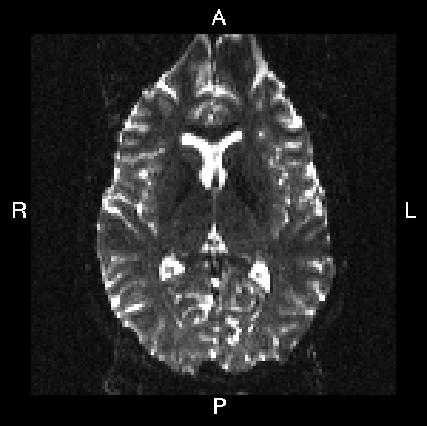
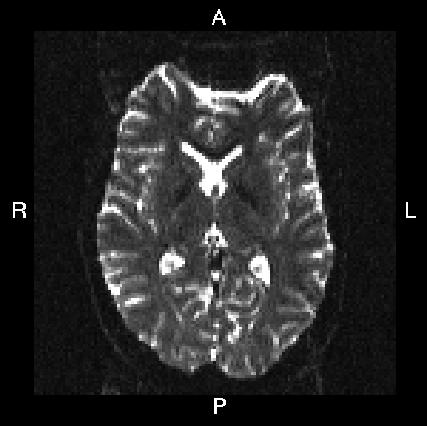
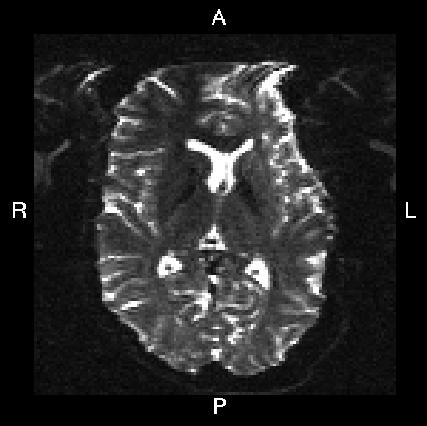
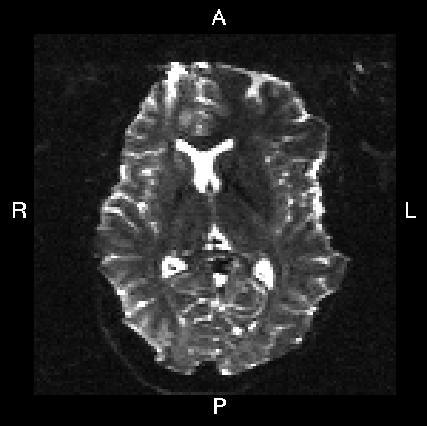
Here is a visual "cheat sheet" for what to put into your --acqp file
| What you see in FSLeyes |
 |
 |
 |
 |
|---|---|---|---|---|
| First three columns in --acqp file | 0 1 0 | 0 -1 0 | 1 0 0 | -1 0 0 |
Do I ever need more than two rows (for eddy) in my --acqp file?
I would say very rarely, if ever.
It was something we suggested in the past, and the rationale for it was to use the movement estimates from the topup output as starting estimates for eddy's movement estimation. Changes to the details of how eddy performs the movement estimation has made that less important. As of the FSL 5.0.9. eddy patch, eddy will start by rigid-body registering all the b=0 and then linearly interpolate the movement parameters from those to the diffusion weighted images. If the b=0 volumes are interspersed among the diffusion images this works really well even in the presence of large subject movements.
If there is a lot of movement in the data, and additional help is still needed one can typically achieve that by adding --niter=8 and --fwhm=10,6,4,2,0,0,0,0 to the eddy command line.
Having said that, there is nothing wrong with having more than two rows in the --acqp file, and it will do no harm.
What does it mean if I have more than two rows in my --acqp file?
It used to be (prior to FSL 5.0.9) that it could be useful to "help" eddy with data with lots of movement. If one had data with several b=0 volumes interleaved into the diffusion acquistion, those b=0 scans could be used for that purpose. It was done by including more than one b=0 volume for one (or both) PE-direction in the --imain file for topup. The relative movement parameters for those was then estimated with very high precision by topup (it is easy to estimate movement between images with identical contrast) and written to the _movpars.txt file. These movement estimates, based on the interleaved b=0 volumes, could then be used by eddy as starting estimates for the harder problem of estimating movements from the dwi volumes.
However, as of FSL 5.0.9, eddy will always start by aligning all the b=0 volumes, and then use those estimates as starting estimates for the dwi alignment. So the motivation for multi-row --acqp files have largely disappeared. Though it is still possible, should one want to.
Will eddy rotate my bvecs for me?
Yes, it will as of the FSL 5.0.9 patch and later versions. As part of the output there will be a file named 'name specified with --out'.eddy_rotated_bvecs. It will have the same format as the file that was supplied as --bvecs, but with the gradient vectors rotated according to the estimated movement parameters. It can be directly supplied to for example dtifit or bedpostx. Note that for the rotation to work the bvecs fed into eddy must be correctly oriented for use by the FSL tools.
What would a good eddy command look like for data with lots of movement?
There are several things we need to take into consideration when there is lots of movement:
- We may need to change the way we run eddy to make sure it converges. Specifically we may want to apply a bit of smoothing to better condition the registration (N.B. this only affects the estimation, the final corrected images will not have any smoothing) and we may want to increase the number of iterations.
- Movement typically also means signal dropout, so we will want to use outliers detection and replacement (
--repol). - When people make sudden movements the slices in a volume may no longer stack up (c.f. fanning a deck of cards), so one may want to use a slice-to-vol (intra-volume) movement correction.
- If there are large out-of-plane rotations the susceptibility-induced field will change, which means that the field estimated by
topupmight no longer be valid. This can be alleviated by modelling the how the field changes with out-of-plane rotation (--estimate_move_by_susceptibility).
An example of an eddy command that does all of that would be
eddy_cuda --imain=my_data --acqp=acqparams.txt --index=index.txt --mask=my_brain_mask
--bvals=bvals --bvecs=bvecs --topup=my_topup --out=my_hifi_data --niter=8
--fwhm=10,6,4,2,0,0,0,0 --repol --ol_type=both --mporder=8 --s2v_niter=8
--json=my.json
In this example --niter=8 and --fwhm=10,6,4,2,0,0,0,0 instructs eddy to run 8 initial iterations with decreasing amounts of smoothing. During these iterations eddy estimates eddy current-induced fields, inter-volume movement parameters and detects and replaces outliers. The outlier detection is requested with the --repol flag. The --ol_type=both parameter instructs eddy to detect both groups of outliers (in a multi-band acquisition, which we assume this data set is, all slices in an MB-group are affected in the same way by subject movement) and individual outlier slices (pulsatile movement of the brain can cause one slice in an MB-group to be an outlier, but not affect the others).
The --mporder=12 and --s2v_niter=8 parameters tells eddy to run an additional 8 iterations. For the additional iterations the movement model is replaced by a slice-to-vol method with 13 (12+1) degrees of freedom per movement parameter and volume. During these additional iterations the estimates of the eddy current-induced fields and the outliers are also further refined. The parameter --json=my.json specifies a JSON file generated by dcm2niix with information of acquisition order and multi-band structure.
Finally, the --estimate_move_by_susceptibility flag and the --mbs_niter=6 tells eddy to run an additional 6 iterations where it is also estimating how the susceptibility-induced field changes as a function of out-of-plane rottaions.
What if I want to run slice-to-volume but don't have a .json file?
Let us be clear -- the best way to specify the multiband/SMS structure of your data is to use the JSON file generated by dcm2niix. It simply reads the information that the scanner vendor has included in the DICOM files, that doesn't fit into the nifti format, and puts it into a JSON "sidecar". A JSON file is simply a text file with key-value pairs which makes it human readable as well as easily decoded in software. The relevant key for the purpose of knowing the multiband/SMS structure is "SliceTiming". Both Siemens and General Electric provide information in their DICOM headers for dcm2niix to be able to deduce this information. If you data originally came from a Phillips scanner it is possible that the information wasn't present in the DICOM, in which case the "SliceTiming" will be missing from your JSON file.
So, what to do if you have "old" data for which you only have nifti files, or if yous JSON sidecar doesn't have the "SliceTiming" key?
The --slspec file is necessary when using the slice-to-vol movement correction introduced in version 5.0.11. It is used to inform eddy of the temporal order in which the slices of a volume was acquired, and also to specify how multiple slices were grouped in a multi-band (MB) acquisition. In certain cases it is quite easy to work this out, for example in the case of a single band acquisition or in the case of an MB acquisition with an odd number of excitations/groups when using the UMinn MB sequence on a Siemens scanner. But even in the case of a single band acquisition there are some "gotchas" that one needs to be aware of.
Hence, when in doubt it is always safest to use the information in the DICOM headers to create the --slspec file. This can be done either directly from the DICOM files, or via a .json file created by your DICOM->niftii conversion software. If you for example have the following file obtained from a conversion with dcm2niix
Sample JSON file from dcm2niix
you can then create your --slspec file using the following Matlab code
fp = fopen('name_of_my_json_file.json','r');
fcont = fread(fp);
fclose(fp);
cfcont = char(fcont');
i1 = strfind(cfcont,'SliceTiming');
i2 = strfind(cfcont(i1:end),'[');
i3 = strfind(cfcont((i1+i2):end),']');
cslicetimes = cfcont((i1+i2+1):(i1+i2+i3-2));
slicetimes = textscan(cslicetimes,'%f','Delimiter',',');
[sortedslicetimes,sindx] = sort(slicetimes{1});
mb = length(sortedslicetimes)/(sum(diff(sortedslicetimes)~=0)+1);
slspec = reshape(sindx,[mb length(sindx)/mb])'-1;
dlmwrite('my_slspec.txt',slspec,'delimiter',' ','precision','%3d');
No doubt there are more elegant ways of doing it, but you would have to write those yourselves.
I look at my eddy-corrected data, and it looks like there is a mis-registration between the b=0 and the dwi volumes.
First of all it should be said that it is not easy to tell by just eye-balling the data. The high signal from CSF in the b=0 data, along with a quite different distribution of CSF on different sides of the brain tends to "trick the eye". What I recommend doing when checking the alignment is to focus on points of high curvature (typically at the "bottom" of sulci) that can be identified in both b=0 and dwi images.
Having said that, it is indeed possible to have such a mismatch. Inside eddy the dwi:s and the b=0 volumes are registered separately. In the initial version of eddy the first b=0 and the first dwi volume were assumed to be in register (by virtue of having been acquired closely in time) and that was that. Later versions have attempted to be a little more clever about it. As of eddy version 5.0.11 we hope to have sorted this out. There is a long description of that here and also how you can affect how it is done through the --dont_sep_offs_move and --dont_peas flags.