FEAT 1 Practical
This tutorial leads you through a standard single-subject analysis with FEAT. There may be moments when you are waiting for programs to run; during those times take a look at the FEAT manual (in particular go to the User Guide and look at the FEAT in Detail section). We also suggest that you do read it carefully after the course, before using FEAT for analysing your own data.
Contents:
- Example real fmri-fluency dataset
- Perform a full first-level analysis of a single subject in an event-related language experiment.
- Featquery
- Use featquery to interrogate results of the previous analysis and extract ROI measurements.
Example real fmri-fluency dataset
cd ~/fsl_course_data/fmri1/fluency_task
The dataset fmri.nii.gz is from a language experiment. The TR
is 4.2 seconds. The experiment is event-related and has three different types
of events:
- Word-generation events (WG): Here the subject is presented with a noun, say for example "car" and his/her task is to come up with a pertinent verb (for example "drive") and then "think that word in his/her head". The subject was explicitly instructed never to say or even mouth a word to prevent movement artefacts.
- Word-shadowing events (WS): Here the subject is presented with a verb and is instructed to simply "think that word in his/her head".
- Null-events (N): These are events where nothing happens, i.e. the cross-hair remains on the screen and no word is presented. The purpose of these "events" is to supply a baseline against which the other two event types can be compared.
Note that there were no additional "instruction events" as part of the experiment. Each event was "its own instruction" in that the class of the word determines the task. This means that even the "shallow" word-shadowing events contain an element of grammatical decoding.
Within one session, the events were presented at a constant ISI (Inter Stimulus Interval) of 6 seconds. For example, the first 72 seconds (twelve events) in this session may have looked like:
N-WS-N-WS-N-WS-N-WG-N-WS-WG-N
The randomisation of event types was "restricted" in the sense that there was an equal number (24) of each event type. In other words, at any given ISI each type of event was equally likely.
The main question for this experiment was to see if the "deeper" language processing in the word-generation task would yield activations over and above that of the shallower processing in the word-shadowing task. But there are also other interesting questions you can ask of the data. So, let us get started with the analysis.
Feat &
(Type Feat_gui & if you are on a Mac).
Data
Feat starts by displaying the Data tab. Press Select 4D data and select fmri.nii.gz (don't just type "fmri.nii.gz" in the file select popup or you probably won't end up setting the full pathname; use the file-select icon on the right to select the input data).
FEAT now knows how many time points (volumes) you have (106 in this dataset). The GUI will set the TR (time between 3D volumes) using information in the NIFTI file, however this information is not always correct (depending on how the conversion to NIFTI was done). So you should always check that the value for TR is correct after the FMRI data is loaded. For these data it should be 4.2 seconds (you might get 4.199, and that is ok).
The High pass filter cutoff is preset to 100secs. This is chosen to remove the worst of the low frequency trends, and is also long enough to avoid removing the signal of interest. In general you need to ensure that this is not set lower than your maximum stimulation period. For a random-order event-related design there is no clear "stimulation period" so in order to assess what the cutoff should be one needs to analyse the frequency-content of our expected activations (remember that the design matrix embodies what we expect to see in the brain). Leave it at the default for now and we'll come back to it when we have specified the design.
Pre-stats and Stats
Press the Pre-stats tab to look at the preprocessing steps. For this experiment we will change Spatial smoothing FWHM (mm) to 7mm, which is slightly more than we normally recommend. All the other default pre-processing steps are fine for this dataset. Note that we are not performing B0 unwarping here to save time, but you would normally switch it on and set it up as you learnt in the Registration practical.
Setting up the design matrix
Select the Stats tab and press Full model setup to setup the GLM details.
Change the Number of EVs to 2 (we have two conditions to model separately - Word-generation and Word-shadowing).
Setup EV1 (Word-generation): First chose a sensible name like
for example Gen for it and change Basic shape to
Custom (3 column format) and select the file word_generation.txt.
Later (when you wait for FEAT to finish the analysis) we will return to
this file and make sure you understand what is in it. Next set
Convolution to Double-Gamma HRF which corresponds to the HRF
you saw in the talks. Leave the setting for Phase, to the default but
unset Add temporal derivative. We would normally recommend
leaving it set (and we will come back to set it) but in order to obtain a very
simple initial design we will unset it for now.
Setup EV2 (Word-shadowing): Chose a name (for example Shad
and change Basic shape to Custom (3 column format) and this time
select the file word_shadowing.txt. Same as for EV1 set
Convolution to Double-Gamma HRF, unset Add temporal
derivative and leave everything else as the defaults.
Setting up contrasts
Now set up the Contrasts (click on the Contrasts & F-tests tab. Set the Number of contrasts to 5 and enter the following contrasts:
- Name the first contrast Generation. Set the contrast to be
sensitive to the activation in word-generation above and beyond that in
rest.
answer
- Name the second contrast Shadowing. Set the contrast to be
sensitive to the activation in word-shadowing above and beyond that in rest.
answer
- Name the third contrast Mean. Set the contrast to be sensitive to
the mean activation in word-generation and word-shadowing being larger
than in rest.
answer
- Name the fourth contrast Shad > Gen. Set the contrast to be
sensitive to the activation in word-shadowing above and beyond that in
word-generation.
answer
- Name the fifth contrast Gen > Shad. Set the contrast to be
sensitive to the activation in word-generation above and beyond that in
word-shadowing.
answer
Next set up an F-test. Set the Number of F-tests to 1 and select the first two contrasts. This spans both conditions and will show you any areas where there is significant activation by Word-generation AND/OR Word-shadowing. Thus the sixth output colour overlay image produced will show where either generation or shadowing activation (or both) occurs; i.e. it will show both on a single image.
Bonus question: Will the F-test with contrasts 1 and 2, and the F-tests with contrasts 3 and 4 result in the same maps or in different maps? answer
Press View design. Make sure you understand the resulting design matrix. Time goes down the page, with every 10 TRs ticked off on the left. The red bar shows the width of the highpass filter (the amplitude of any signal much longer than it will get reduced). There are 2 columns in the design corresponding to our predictions about BOLD activity from Word-generation and Word-shadowing respectively. Hopefully they will be familiar from the talk you have just heard. The contrasts appear at the bottom of the image, with the F-test to the right of the contrasts. Note that you can make the design matrix display disappear just by clicking on it once. For now, leave the design matrix display up (Press View design again if necessary).
Temporal derivatives
Now, return to the EVs tab and FOR BOTH EVs select Add temporal derivative.
Press View design again. You now see 4 columns with columns 1 and 3 being the same as before and column 2 and 4 being "new". These are the temporal derivatives that are used to correct for timing errors caused either by slight experimental errors in synchronising the times of the scanner with the stimulus presentation and/or inter-subject differences in the delay inherent in the HRF. Now press "Done" and dismiss the view of the design matrix.
High pass filtering
Do you remember that we said we should return to the issue of High pass filtering once we knew the design (and with that the expected frequency content of the signal we expect/hope to see)? Now the time has come. Press the Data-tab to make sure that High pass filter cutoff (s) is set to 100. Next press the Misc-tab where there will be a button saying Estimate High Pass Filter. Press this button and then go back to the Data-tab to see what has happened. This should now have changed to 90 seconds. FSL has calculated this for you by analysing the frequency content of the design and then selected a cutoff so that 90% of our expected signal is still in the data after filtering. (N.B. that it is just a fluke that 90% happened to translate into 90 seconds in this particular case)
Look at the Post-stats section - the defaults are fine; cluster-based thresholding will be carried out.
Registration
Select the Registration tab. By default FEAT will
register the middle-timepoint FMRI image (saved as
example_func in the .feat output directory)
to the standard space template. We recommend in general turning on the
Main structural image option so that the lowres FMRI image is first
registered to a brain-extracted highres structural image from the same
subject; this highres image is then registered to the standard
space template, and then the two registrations are combined to give an
example_func2standard.mat transform which can be used later to
resample the FMRI stats into standard space.
Set the Main structural image file
to structural_brain.nii.gz and change BBR to 7 DOF (note that we have already
run BET on this, and in order to save time in this practical session we are
not using the BBR method, but we strongly recommend that BBR is used
generally). Leave Standard space turned on
with MNI152_T1_2mm_brain.nii.gz selected and set the DOF to
12. Instead of the linear (12 DOF) registration we are using here, the more accurate nonlinear registration is
normally recommended for registration to MNI space by selecting the
"nonlinear" option, but we use linear here as it is faster for this
practical.
Go!
You are now ready to run FEAT. Press Go. A web browser should appear, and as FEAT completes the different stages of processing, you will see messages appear in the Log section. Whether the web browser (and indeed the FEAT GUI) is left displayed or is closed, FEAT will continue to run in the background. For now, leave the web browser open so that you can monitor FEAT's progress. FEAT will take 2-5 minutes to complete.
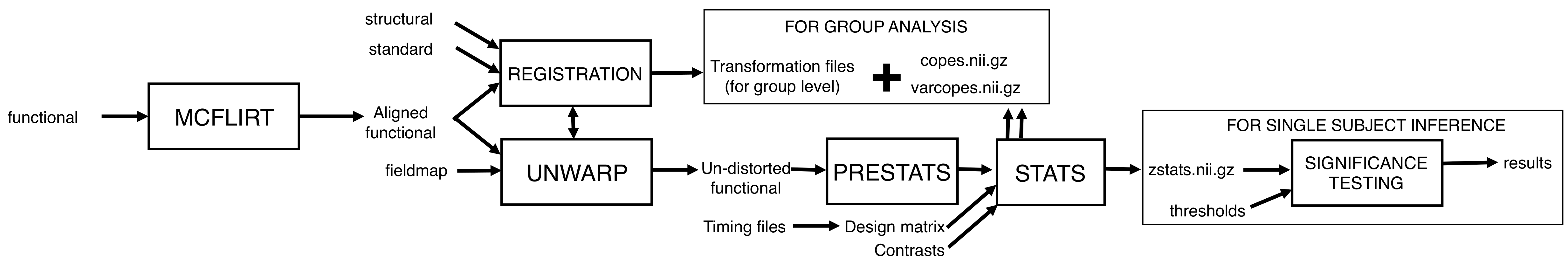
While you wait
Take a look at the flow diagram below which summarises all of the steps that you just set up in Feat. Hopefully it will help you to see the big picture, and avoid getting bogged down in the details:
Whilst FEAT is running, run FSLeyes to have a quick look at the
different images mentioned above: start by looking at
structural_brain.nii.gz, and then view
fmri.nii.gz. Note that when viewing the 4D image you can see the
image time series as a movie by pressing on the movie icon
(![]() ), and you can also see time series
plots by pressing View > Timeseries.
), and you can also see time series
plots by pressing View > Timeseries.
Create a mask to probe statistics
Whilst FEAT is still running, we will now use FSLeyes to create a mask in standard space that will be used later to find out about activation statistics from within the mask. An alternative method to create a mask was explained in the Registration practical.
- Reopen FSLeyes and load the standard space template image
$FSLDIR/data/standard/MNI152_T1_2mm($FSLDIRis an environment variable indicating the directory in which FSL is installed, you can typeecho $FSLDIRto see what this is set to). Inside FSLeyes you can use the File -> Add standard menu option to find these standard space images quickly. - Open the atlas panel via Settings -> Ortho View 1 -> Atlases, and enable the Harvard-Oxford cortical, Harvard-Oxford subcortical and Juelich Histological atlases. Move the cursor around a little in the standard brain and see how the labels and numbers in the atlas tool window changes. If you have a favourite part of the brain and you happen to know where it is you can move the cursor there and see if the atlas tool agrees with you.
- Now select the Atlas search tab in the atlas panel, and choose the Juelich Histological Atlas from the list on the left (just click on the text; do not tick the check box). You will now see a list of all the structures in that atlas on the right.
- Type
ba4in the text box above the structure list to filter the structures that are shown. Click the check boxes next to GM Broca's area BA44 L and GM Broca's area BA45 L. - You will notice that in the FSLeyes overlay list, two images have been added with name corresponding to the regions you just selected. If you select one of those and then move the cursor around you will notice its intensity values in changing between 0 and 100. These values reflect the probability that a given voxel (cursor position) is indeed part of that structure.
- Press the save icon
(
 )
next to the image juelich/prob/GM Broca's area BA44 L in the overlay list and
save the image to a file called
)
next to the image juelich/prob/GM Broca's area BA44 L in the overlay list and
save the image to a file called BA44. Repeat this process for BA45. What we will do next is to create a mask which has the value 1 for each voxel that has a 50% or greater chance of belonging to BA44 and/or BA45. We do this by typing (in the terminal window):
fslmaths BA44 -add BA45 -thr 50 -bin Broca
The end result of this is a file named
Broca.nii.gzthat we will later use as a mask to plot time-series of our results. If you want to convince yourself that this file indeed contains what it should you can typefsleyes -std Broca -cm red
and have a look.
Look at the EV specification
Lastly, whilst FEAT is running take a look at the files that we used to
specify our design, i.e. word_generation.txt
and word_shadowing.txt. We do this by typing (still in the
terminal window):
cat word_generation.txt
The cat command will show you the contents of the file.
Once you've finished looking at word_generation.txt,
run cat word_shadowing.txt to look at the timing
information for the word shadowing task.
While FEAT is running it will display STILL RUNNING in the main FEAT report page, which is replaced by Finished at ... when it is done. Once FEAT has finished, look carefully at the various sections of the web page report, including motion correction plots in the Pre-stats section, the colour-rendered activation images and timeseries plots in the Post-stats section, and the Registration results. Note that if you click on the activation images you get a table of cluster co-ordinates.
Featquery
Featquery allows you to calculate certain data statistics, either at a
voxel of interest, or averaged over a region of interest using a mask. We will
use the standard-space mask which we created earlier. Start
up Featquery from the terminal:
Featquery &
(or Featquery_gui & if you are on a Mac).
Select the fmri.feat directory created by your first analysis
on the fmri-fluency dataset.
Featquery automatically reads the FEAT directory and gives you the appropriate options as to which statistics you can choose to investigate.
- Select the following statistics for the contrast that looks at activation
for word-generation (i.e. the [1 0] contrast), the contrast
that looks at activation for word-shadowing (i.e. the [0 1]
contrast) and the contrast that looks at activation for word-generation over
and above word-shadowing (i.e. the [1 -1]
contrast), i.e. contrasts number 1, 2 and 5:
- stats/cope (unthresholded contrast of parameter estimate)
- stats/tstat (unthresholded t statistics)
- stats/zstat (unthresholded z statistics)
- thresh_zstat (thresholded z statistics)
- In the Input ROI selection panel, enter the mask that you created
earlier (i.e.
Broca.nii.gz) as the Mask Image (note - Featquery can take either a standard-space mask OR a lowres one in the original dataspace OR a mask in the space of the structural image) - In the Output options section, select the Convert PE/COPE values to % option
- Select the Do not binarise mask (allow weighting) option
- Select an atlas (inside the Output options section), for example the Harvard-Oxford Cortical Structural Atlas. The local maxima voxels reported by Featquery will be related to structures in the selected atlas.
- Press Go and a web browser showing the estimated statistics should popup shortly (possibly after a minute or two).
The resulting web page will contain a table summarizing each of the statistics that you asked Featquery to report on in step 1. The first column gives the statistic name. The second column gives the number of non-zero voxels in the mask. The next group of columns gives a summary of the distribution of values within the mask. Finally, the last group of columns contains the position of the maximum in voxel space, in mm space, and in the atlas space selected in step 5. Plots of the timeseries at the maximum z-stats are available by clicking the link labeled "Masked time series plot" just below the image of the mask at the top of the page.
The End.